Tutorial 1: Data Frames (Wrangling an Image Dataset)
Contents
Tutorial 1: Data Frames (Wrangling an Image Dataset)¶
In this tutorial, we’ll learn how to use Meerkat to explore a dataset of images. We’ll use the imagenette dataset, a small 10-class subset of ImageNet.
Through this tutorial, you will learn about:
the concept of data frames in Meerkat
the different types of data that you can store in a Meerkat data frame
how to use data frames to explore unstructured data
The content of this tutorial is provided in the tutorial-0.ipynb Jupyter notebook.
🔮 Importing Meerkat¶
Let’s start by importing Meerkat.
import os
import meerkat as mk
import numpy as np
💾 Downloading the data¶
First, we’ll download some data to explore. We’re going to use the Imagenette dataset, a small subset of the original ImageNet. This dataset is made up of 10 classes (e.g. “garbage truck”, “gas pump”, “golf ball”).
Download time: <1 minute
Download size: 130M
In addition to downloading the data, download_imagnette prepares a CSV, imagenette.csv, with a row for each image.
from meerkat.datasets.imagenette import download_imagenette
dataset_dir = "./downloads"
os.makedirs(dataset_dir, exist_ok=True)
download_imagenette(dataset_dir, overwrite=True);
Extracting tar archive, this may take a few minutes...
Let’s take a look at the CSV.
!head -n 5 downloads/imagenette2-160/imagenette.csv
label,split,img_path
cassette player,train,train/n02979186/n02979186_9036.JPEG
cassette player,train,train/n02979186/n02979186_11957.JPEG
cassette player,train,train/n02979186/n02979186_9715.JPEG
cassette player,train,train/n02979186/n02979186_21736.JPEG
Next, we’ll load it into a Meerkat DataFrame.
📸 Creating an image DataFrame¶
For more information on creating DataFrames from various data sources, see the user guide section on I/O.
Meerkat’s core contribution is the DataFrame, a simple columnar data abstraction. The Meerkat DataFrame can house columns of arbitrary type – from integers and strings to complex, high-dimensional objects like videos, images, medical volumes and graphs.
We’re going to build a DataFrame out of the imagenette.csv file from the download above.
# Create a `DataFrame`
df = mk.from_csv("./downloads/imagenette2-160/imagenette.csv")
# Create an `ImageColumn`` and add it to the `DataFrame`
df["img"] = mk.image(
df["img_path"],
base_dir=os.path.join(dataset_dir, "imagenette2-160")
)
df
The call to head shows the first few rows in the DataFrame. You can see that there are a few metadata columns, as well as the “img” column we added in.
🗂 Selecting data¶
For more information see the user guide section on Data Selection.
When we create an ImageColumn we don’t load the images into memory. Instead, ImageColumn keeps track of all those filepaths we passed in and only loads the images when they are needed.
When we select a row of the ImageColumn, we get an instance FileCell back. A FileCell is an object that holds everything we need to materialize the cell (e.g. the filepath to the image and the loading function), but stops just short of doing so.
img_cell = df["img"][0]
print(f"Indexing the `ImageColumn` returns an object of type: {type(img_cell)}.")
Indexing the `ImageColumn` returns an object of type: <class 'meerkat.columns.deferred.file.FileCell'>.
To actually materialize the image, we simply call the cell.
img = img_cell()
img

We can subselect a batch of images by indexing with a slice. Notice that this returns a smaller DataFrame.
imgs = df["img"][1:4]
print(f"Indexing a slice of the `ImageColumn` returns a: {type(imgs)}.")
imgs
Indexing a slice of the `ImageColumn` returns a: <class 'meerkat.columns.deferred.file.FileColumn'>.
| (FileColumn) | |
|---|---|
| 0 | |
| 1 | |
| 2 |
The whole batch of images can be loaded together by calling the column.
imgs();
One can load multiple rows using any one of following indexing schemes:
Slice indexing: e.g.
column[4:10]Integer array indexing: e.g.
column[[0, 4, 6, 11]]Boolean array indexing: e.g.
column[np.array([True, False, False ..., True, False])]
📎 Aside: ImageColumn under the hood, DeferredColumn.¶
If you check out the implementation of ImageColumn (at meerkat/column/image_column.py), you’ll notice that it’s a super simple subclass of DeferredColumn.
What’s a DeferredColumn?
In meerkat, high-dimensional data types like images and videos are typically stored in a DeferredColumn. A DeferredColumn wraps around another column and lazily applies a function to it’s content as it is indexed. Consider the following example, where we create a simple meerkat column…
col = mk.column([0,1,2])
…and wrap it in a deferred column.
dcol = col.defer(fn=lambda x: x + 10)
dcol[1]() # the function is only called at this point!
Critically, the function inside a lambda column is only called at the time the column is called! This is very useful for columns with large data types that we don’t want to load all into memory at once. For example, we could create a DeferredColumn that lazily loads images…
>>> filepath_col = mk.PandasSeriesColumn(["path/to/image0.jpg", ...])
>>> img_col = filepath.defer(lambda x: load_image(x))
An ImageColumn is a just a DeferredColumn like this one, with a few more bells and whistles!
🛠 Applying operations over the DataFrame.¶
When analyzing data, we often perform operations on each example in our dataset (e.g. compute a model’s prediction on each example, tokenize each sentence, compute a model’s embedding for each example) and store them. The DataFrame makes it easy to perform these operations:
Produce new columns (via
DataFrame.map)Produce new columns and store the columns alongside the original data (via
DataFrame.update)Extract an important subset of the datset (via
DataFrame.filter).
Under the hood, dataloading is multiprocessed so that costly I/O doesn’t bottleneck our computation.
Let’s start by filtering the DataFrame down to the examples in the validation set.
valid_df = df.filter(lambda split: split == "valid", is_batched_fn=True, batch_size=len(df))
🫐 Using DataFrame.map to compute average intensity of the blue color channel in the images.¶
To demonstrate the utility of the map operation, we’ll explore the relationship between the “blueness” of an image and the class of the image.
We’ll quantify the “blueness” of each image by simply computing the mean intensity of the blue color channel. This can be accomplished with a simple map operation over the DataFrame:
blue_col = valid_df.map(
lambda img: np.array(img)[:, :, 2].mean(),
num_workers=2
)
# Add the intensities as a new column in the `DataFrame`
valid_df["avg_blue"] = blue_col
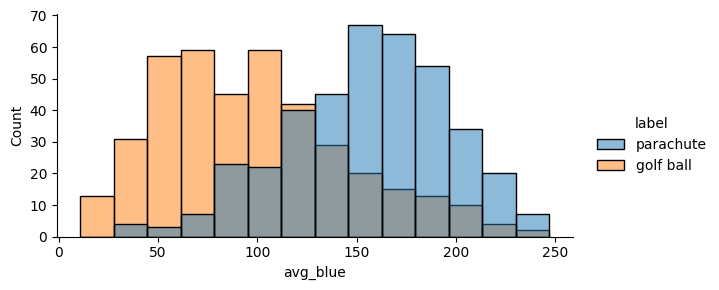
🪂 vs. ⛳️¶
Next, we’ll explore the relationship between blueness and the class label of the image. To do so, we’ll compare the blue intensity distribution of images labeled “parachute” to the distribution of of images labeled “golf ball”.
Using the seaborn plotting package and our DataFrame, this can be accomplished in one line:
## OPTIONAL: this cell requires the seaborn dependency: https://seaborn.pydata.org/installing.html
import seaborn as sns
plot_df = valid_df[np.isin(valid_df["label"], ["golf ball", "parachute"])]
sns.displot(
data=plot_df.to_pandas(),
x="avg_blue",
hue="label",
height=3,
aspect=2
)
/home/runner/work/meerkat/meerkat/meerkat/dataframe.py:901: UserWarning: Could not convert column img of type <class 'meerkat.columns.deferred.file.FileColumn'>, it will be dropped from the output.
warnings.warn(
<seaborn.axisgrid.FacetGrid at 0x7faaf89b0970>
valid_df["img"][int(np.argmax(valid_df["avg_blue"]))]()

📉 ML with images in meerkat.¶
Let’s do some machine learning on our Imagenette DataFrame.
We’ll take a resnet18 pretrained on the full ImageNet dataset, perform inference on the validation set, and analyze the model’s predictions and activations.
The cell below downloads the model..
import torch
from torchvision.models import resnet18
import torchvision.transforms as transforms
model = resnet18(weights="ResNet18_Weights.IMAGENET1K_V1")
Downloading: "https://download.pytorch.org/models/resnet18-f37072fd.pth" to /home/runner/.cache/torch/hub/checkpoints/resnet18-f37072fd.pth
0%| | 0.00/44.7M [00:00<?, ?B/s]
37%|███▋ | 16.7M/44.7M [00:00<00:00, 174MB/s]
100%|██████████| 44.7M/44.7M [00:00<00:00, 239MB/s]
💈 Applying a transform to the images.¶
In order to do inference, we’ll need to create a new DeferredColumn. The ImageColumn we defined above (i.e. "img_path"), does not apply any transforms after loading and simply returns a PIL image. Before passing the images through the model, we need to convert the PIL image to a torch.Tensor and normalize the color channels (along with a few other transformations).
Note: the transforms defined below are the same as the ones used by torchvision, see here.
In the cell below, we specify a transform when creating the ImageColumn.
# Define transform
transform = transforms.Compose([
transforms.Resize(256),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize(
mean=[0.485, 0.456, 0.406],
std=[0.229, 0.224, 0.225]),
])
# Create new column with transform
valid_df["input"] = valid_df["img"].defer(transform)
Notice that indexing this new column returns a torch.Tensor, not a PIL image…
img = valid_df["input"][0]()
print(f"Indexing the `ImageColumn` returns an object of type: {type(img)}.")
Indexing the `ImageColumn` returns an object of type: <class 'torch.Tensor'>.
… and that indexing a slice of this new column returns a TensorColumn.
col = img = valid_df["input"][:3]()
print(f"Indexing a slice of the `ImageColumn` returns an object of type: {type(img)}.")
col
Indexing a slice of the `ImageColumn` returns an object of type: <class 'meerkat.columns.tensor.torch.TorchTensorColumn'>.
| (TorchTensorColumn) | |
|---|---|
| 0 | torch.Tensor(shape=torch.Size([3, 224, 224])) |
| 1 | torch.Tensor(shape=torch.Size([3, 224, 224])) |
| 2 | torch.Tensor(shape=torch.Size([3, 224, 224])) |
Let’s see what the full DataFrame looks like now.
valid_df.head()
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
File /opt/hostedtoolcache/Python/3.9.17/x64/lib/python3.9/site-packages/IPython/core/formatters.py:922, in IPythonDisplayFormatter.__call__(self, obj)
920 method = get_real_method(obj, self.print_method)
921 if method is not None:
--> 922 method()
923 return True
File ~/work/meerkat/meerkat/meerkat/interactive/graph/marking.py:74, in unmarked.__call__.<locals>.decorate_context(*args, **kwargs)
71 @wraps(func)
72 def decorate_context(*args, **kwargs):
73 with self.clone():
---> 74 return reactive(func, nested_return=False)(*args, **kwargs)
File ~/work/meerkat/meerkat/meerkat/interactive/graph/reactivity.py:211, in reactive.<locals>.__reactive.<locals>.wrapper(*args, **kwargs)
209 # Call the function on the args and kwargs
210 with unmarked():
--> 211 result = fn(*unpacked_args, **unpacked_kwargs)
213 # TODO: Check if result is equal to one of the inputs.
214 # If it is, we need to copy it.
216 if _is_unmarked_context or _force_no_react or not any_inputs_marked:
217 # If we are in an unmarked context, then we don't need to create
218 # any nodes in the graph.
219 # `fn` should be run as normal.
File ~/work/meerkat/meerkat/meerkat/dataframe.py:193, in DataFrame._ipython_display_(self)
190 max_rows = meerkat.config.display.max_rows
191 df, formatters = self._repr_pandas_(max_rows=max_rows)
192 return display(
--> 193 HTML(df.to_html(formatters=formatters, max_rows=max_rows, escape=False))
194 )
196 return self.gui.table()._ipython_display_()
File /opt/hostedtoolcache/Python/3.9.17/x64/lib/python3.9/site-packages/pandas/core/frame.py:3129, in DataFrame.to_html(self, buf, columns, col_space, header, index, na_rep, formatters, float_format, sparsify, index_names, justify, max_rows, max_cols, show_dimensions, decimal, bold_rows, classes, escape, notebook, border, table_id, render_links, encoding)
3109 formatter = fmt.DataFrameFormatter(
3110 self,
3111 columns=columns,
(...)
3126 show_dimensions=show_dimensions,
3127 )
3128 # TODO: a generic formatter wld b in DataFrameFormatter
-> 3129 return fmt.DataFrameRenderer(formatter).to_html(
3130 buf=buf,
3131 classes=classes,
3132 notebook=notebook,
3133 border=border,
3134 encoding=encoding,
3135 table_id=table_id,
3136 render_links=render_links,
3137 )
File /opt/hostedtoolcache/Python/3.9.17/x64/lib/python3.9/site-packages/pandas/io/formats/format.py:1108, in DataFrameRenderer.to_html(self, buf, encoding, classes, notebook, border, table_id, render_links)
1099 Klass = NotebookFormatter if notebook else HTMLFormatter
1101 html_formatter = Klass(
1102 self.fmt,
1103 classes=classes,
(...)
1106 render_links=render_links,
1107 )
-> 1108 string = html_formatter.to_string()
1109 return save_to_buffer(string, buf=buf, encoding=encoding)
File /opt/hostedtoolcache/Python/3.9.17/x64/lib/python3.9/site-packages/pandas/io/formats/html.py:77, in HTMLFormatter.to_string(self)
76 def to_string(self) -> str:
---> 77 lines = self.render()
78 if any(isinstance(x, str) for x in lines):
79 lines = [str(x) for x in lines]
File /opt/hostedtoolcache/Python/3.9.17/x64/lib/python3.9/site-packages/pandas/io/formats/html.py:83, in HTMLFormatter.render(self)
82 def render(self) -> list[str]:
---> 83 self._write_table()
85 if self.should_show_dimensions:
86 by = chr(215) # ×
File /opt/hostedtoolcache/Python/3.9.17/x64/lib/python3.9/site-packages/pandas/io/formats/html.py:258, in HTMLFormatter._write_table(self, indent)
255 if self.fmt.header or self.show_row_idx_names:
256 self._write_header(indent + self.indent_delta)
--> 258 self._write_body(indent + self.indent_delta)
260 self.write("</table>", indent)
File /opt/hostedtoolcache/Python/3.9.17/x64/lib/python3.9/site-packages/pandas/io/formats/html.py:406, in HTMLFormatter._write_body(self, indent)
404 def _write_body(self, indent: int) -> None:
405 self.write("<tbody>", indent)
--> 406 fmt_values = self._get_formatted_values()
408 # write values
409 if self.fmt.index and isinstance(self.frame.index, MultiIndex):
File /opt/hostedtoolcache/Python/3.9.17/x64/lib/python3.9/site-packages/pandas/io/formats/html.py:401, in HTMLFormatter._get_formatted_values(self)
399 def _get_formatted_values(self) -> dict[int, list[str]]:
400 with option_context("display.max_colwidth", None):
--> 401 fmt_values = {i: self.fmt.format_col(i) for i in range(self.ncols)}
402 return fmt_values
File /opt/hostedtoolcache/Python/3.9.17/x64/lib/python3.9/site-packages/pandas/io/formats/html.py:401, in <dictcomp>(.0)
399 def _get_formatted_values(self) -> dict[int, list[str]]:
400 with option_context("display.max_colwidth", None):
--> 401 fmt_values = {i: self.fmt.format_col(i) for i in range(self.ncols)}
402 return fmt_values
File /opt/hostedtoolcache/Python/3.9.17/x64/lib/python3.9/site-packages/pandas/io/formats/format.py:895, in DataFrameFormatter.format_col(self, i)
893 frame = self.tr_frame
894 formatter = self._get_formatter(i)
--> 895 return format_array(
896 frame.iloc[:, i]._values,
897 formatter,
898 float_format=self.float_format,
899 na_rep=self.na_rep,
900 space=self.col_space.get(frame.columns[i]),
901 decimal=self.decimal,
902 leading_space=self.index,
903 )
File /opt/hostedtoolcache/Python/3.9.17/x64/lib/python3.9/site-packages/pandas/io/formats/format.py:1330, in format_array(values, formatter, float_format, na_rep, digits, space, justify, decimal, leading_space, quoting, fallback_formatter)
1314 digits = get_option("display.precision")
1316 fmt_obj = fmt_klass(
1317 values,
1318 digits=digits,
(...)
1327 fallback_formatter=fallback_formatter,
1328 )
-> 1330 return fmt_obj.get_result()
File /opt/hostedtoolcache/Python/3.9.17/x64/lib/python3.9/site-packages/pandas/io/formats/format.py:1363, in GenericArrayFormatter.get_result(self)
1362 def get_result(self) -> list[str]:
-> 1363 fmt_values = self._format_strings()
1364 return _make_fixed_width(fmt_values, self.justify)
File /opt/hostedtoolcache/Python/3.9.17/x64/lib/python3.9/site-packages/pandas/io/formats/format.py:1430, in GenericArrayFormatter._format_strings(self)
1428 for i, v in enumerate(vals):
1429 if not is_float_type[i] and leading_space or self.formatter is not None:
-> 1430 fmt_values.append(f" {_format(v)}")
1431 elif is_float_type[i]:
1432 fmt_values.append(float_format(v))
File /opt/hostedtoolcache/Python/3.9.17/x64/lib/python3.9/site-packages/pandas/io/formats/format.py:1410, in GenericArrayFormatter._format_strings.<locals>._format(x)
1407 return repr(x)
1408 else:
1409 # object dtype
-> 1410 return str(formatter(x))
File ~/work/meerkat/meerkat/meerkat/interactive/formatter/base.py:351, in DeferredFormatter.html(self, cell)
350 def html(self, cell: DeferredCell):
--> 351 return self.wrapped.html(cell())
File ~/work/meerkat/meerkat/meerkat/interactive/formatter/base.py:351, in DeferredFormatter.html(self, cell)
350 def html(self, cell: DeferredCell):
--> 351 return self.wrapped.html(cell())
TypeError: 'Tensor' object is not callable
DataFrame(nrows: 5, ncols: 6)
💫 Computing model predictions and activations.¶
We’d like to perform inference and extract:
Output predictions
Output class probabilities
Model activations
Note: in order to extract model activations, we’ll need to use a PyTorch forward hook and register it on the final layer of the ResNet. Forward hooks are just functions that get executed on the forward pass of a torch.nn.Module.
# Define forward hook in ActivationExtractor class
class ActivationExtractor:
"""Extracting activations a targetted intermediate layer"""
def __init__(self):
self.activation = None
def forward_hook(self, module, input, output):
self.activation = output
# Register forward hook
extractor = ActivationExtractor()
model.layer4.register_forward_hook(extractor.forward_hook);
We want to apply a forward pass to each image in the DataFrame and store the outputs as new columns: DataFrame.map is perfectly suited for this task.
# 1. Move the model to GPU, if available
# device = 0
device = "cpu"
model.to(device).eval()
# 2. Define a function that runs a forward pass over a batch
@torch.no_grad()
def predict(input: mk.TensorColumn):
x: torch.Tensor = input.data.to(device) # We get the underlying torch tensor with `data` and move to GPU
out: torch.Tensor = model(x) # Run forward pass
# Return a dictionary with one key for each of the new columns. Each value in the
# dictionary should have the same length as the batch.
return {
"pred": out.cpu().numpy().argmax(axis=-1),
"probs": torch.softmax(out, axis=-1).cpu(),
"activation": extractor.activation.mean(dim=[-1,-2]).cpu()
}
# 3. Apply the update. Note that the `predict` function operates on batches, so we set
# `batched=True`. Also, the `predict` function only accesses the "input" column, by
# specifying that here we instruct update to only load that one column and skip others
pred_df = valid_df.map(function=predict, is_batched_fn=True, batch_size=32)
valid_df = mk.concat([valid_df, pred_df], axis="columns")
The predictions, output probabilities, and activations are now stored alongside the examples in the DataFrame.
valid_df[["label_id", "input", "pred", "probs", "activation"]].head()
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
Cell In[20], line 1
----> 1 valid_df[["label_id", "input", "pred", "probs", "activation"]].head()
File ~/work/meerkat/meerkat/meerkat/interactive/graph/reactivity.py:211, in reactive.<locals>.__reactive.<locals>.wrapper(*args, **kwargs)
209 # Call the function on the args and kwargs
210 with unmarked():
--> 211 result = fn(*unpacked_args, **unpacked_kwargs)
213 # TODO: Check if result is equal to one of the inputs.
214 # If it is, we need to copy it.
216 if _is_unmarked_context or _force_no_react or not any_inputs_marked:
217 # If we are in an unmarked context, then we don't need to create
218 # any nodes in the graph.
219 # `fn` should be run as normal.
File ~/work/meerkat/meerkat/meerkat/dataframe.py:537, in DataFrame.__getitem__(self, posidx)
535 @reactive()
536 def __getitem__(self, posidx):
--> 537 return self._get(posidx, materialize=False)
File ~/work/meerkat/meerkat/meerkat/interactive/graph/marking.py:74, in unmarked.__call__.<locals>.decorate_context(*args, **kwargs)
71 @wraps(func)
72 def decorate_context(*args, **kwargs):
73 with self.clone():
---> 74 return reactive(func, nested_return=False)(*args, **kwargs)
File ~/work/meerkat/meerkat/meerkat/interactive/graph/reactivity.py:211, in reactive.<locals>.__reactive.<locals>.wrapper(*args, **kwargs)
209 # Call the function on the args and kwargs
210 with unmarked():
--> 211 result = fn(*unpacked_args, **unpacked_kwargs)
213 # TODO: Check if result is equal to one of the inputs.
214 # If it is, we need to copy it.
216 if _is_unmarked_context or _force_no_react or not any_inputs_marked:
217 # If we are in an unmarked context, then we don't need to create
218 # any nodes in the graph.
219 # `fn` should be run as normal.
File ~/work/meerkat/meerkat/meerkat/interactive/graph/reactivity.py:144, in reactive.<locals>.__reactive.<locals>.wrapper.<locals>._fn_outer_wrapper.<locals>._fn_wrapper(*args, **kwargs)
142 @wraps(_fn)
143 def _fn_wrapper(*args, **kwargs):
--> 144 return _fn(*args, **kwargs)
File ~/work/meerkat/meerkat/meerkat/dataframe.py:525, in DataFrame._get(self, posidx, materialize)
523 if not set(posidx).issubset(self.columns):
524 missing_cols = set(posidx) - set(self.columns)
--> 525 raise KeyError(f"DataFrame does not have columns {missing_cols}")
527 df = self._clone(data=self.data[posidx])
528 return df
KeyError: "DataFrame does not have columns {'label_id'}"
🎯 Computing metrics and analyzing performance.¶
Computing statistics on Meerkat DataFrames is straightforward because standard NumPy operators and functions can be applied directly to a NumpyArrayColumn. We take advantage of this below to compute the accuracy of the model.
valid_df["correct"] = valid_df["pred"] == valid_df["label_idx"].data
accuracy = valid_df["correct"].mean()
print(f"Micro accuracy across the ten Imagenette classes: {accuracy:0.3}")
Furthermore, since the DataFrame is naturally converted to a Pandas DataFrame, it’s easy to use data visualization tools that interface with Pandas (e.g. seaborn, bokeh).
## OPTIONAL: this cell requires the seaborn dependency: https://seaborn.pydata.org/installing.html
import seaborn as sns
import matplotlib.pyplot as plt
sns.barplot(data=valid_df.to_pandas(), y="label", x="correct");
🔎 Exploring model activations.¶
To better understand the behavior of our model, we’ll explore the activations of the final convolutional layer of the ResNet. Recall that when we performed our forward pass, we extracted these activations and stored them in a new column called "activation".
Unlike the the NumpyArrayColumns we’ve been working with so far, the activation column has an additional dimension of size 512.
To visualize the activations, we’ll use a dimensionality reduction technique (UMAP) to embed the activations in two dimensions. We’ll store these embeddings in two new columns “umap_0” and “umap_1”.
## OPTIONAL: this cell requires the umap dependency: https://umap-learn.readthedocs.io/en/latest/
!pip install umap-learn
from umap import UMAP
# 1. Compute UMAP embedding
reducer = UMAP()
embs = reducer.fit_transform(valid_df["activation"])
# 2. Add the embedding to the DataFrame as two new columns
valid_df["umap_0"] = embs[:, 0]
valid_df["umap_1"] = embs[:, 1]
## OPTIONAL: this cell requires the seaborn dependency: https://seaborn.pydata.org/installing.html
import seaborn as sns
import matplotlib.pyplot as plt
sns.scatterplot(data=valid_df.to_pandas(), x="umap_0", y="umap_1", hue="label");
plt.legend(bbox_to_anchor=(1.05, 1), loc='upper left')
💾 Writing a DataFrame to disk.¶
Finally, we can write the updated DataFrame, with all the activations and predictions included, to disk for later use.
DataFrame size on disk: 25MB
On disk, the DataFrame is stored in a directory at the path passed to DataFrame.write. Within that directory, each column is stored separately. This allows us to read only a subset of columns from DataFrame on disk. Use the file explorer to the left to further expore the file structure of the DataFrame.
valid_df
| +-- meta.yml
| +-- state.dill
| +-- columns
| +-- activation
| +-- avg_blue
| +-- correct
| ...
| +-- umap_1
valid_df.write(os.path.join(dataset_dir, "valid_df"))
valid_df = mk.read(os.path.join(dataset_dir, "valid_df"))

