Quickstart: Data Frames
Contents
Quickstart: Data Frames¶
This quickstart provides a quick walkthrough of the Meerkat data frame, which allows users to interact with unstructured data alongside standard tabular data.
import os
import meerkat as mk
import numpy as np
💾 Downloading the data¶
First, we’ll download some data to explore. We’re going to use the Imagenette dataset, a small subset of the original ImageNet. This dataset is made up of 10 classes (e.g. “garbage truck”, “gas pump”, “golf ball”).
Download time: < 1 minute
Download size: 130M
In addition to downloading the data, download_imagnette prepares a CSV, imagenette.csv, with a row for each image.
from meerkat.datasets.imagenette import download_imagenette
dataset_dir = "./downloads"
os.makedirs(dataset_dir, exist_ok=True)
download_imagenette(dataset_dir, overwrite=True)
Extracting tar archive, this may take a few minutes...
Let’s take a look at the CSV.
!head -n 5 downloads/imagenette2-160/imagenette.csv
label,split,img_path
cassette player,train,train/n02979186/n02979186_9036.JPEG
cassette player,train,train/n02979186/n02979186_11957.JPEG
cassette player,train,train/n02979186/n02979186_9715.JPEG
cassette player,train,train/n02979186/n02979186_21736.JPEG
Next, we’ll load it into a Meerkat DataFrame.
📸 Creating an image DataFrame¶
For more information on creating DataFrames from various data sources, see the user guide section on I/O.
Meerkat’s core contribution is the DataFrame, a simple columnar data abstraction. The Meerkat DataFrame can house columns of arbitrary type – from integers and strings to complex, high-dimensional objects like videos, images, medical volumes and graphs.
We’re going to build a DataFrame out of the imagenette.csv file from the download above.
# Create a `DataFrame`
df = mk.from_csv("./downloads/imagenette2-160/imagenette.csv")
# Create an `ImageColumn`` and add it to the `DataFrame`
df["img"] = mk.image(
df["img_path"],
base_dir=os.path.join(dataset_dir, "imagenette2-160")
)
df
| label | split | img_path | img | |
|---|---|---|---|---|
| 0 | cassette player | train | train/n02979186/n02979186_9036.JPEG | |
| 1 | cassette player | train | train/n02979186/n02979186_11957.JPEG | |
| 2 | cassette player | train | train/n02979186/n02979186_9715.JPEG | |
| 3 | cassette player | train | train/n02979186/n02979186_21736.JPEG | |
| 4 | cassette player | train | train/n02979186/ILSVRC2012_val_00046953.JPEG | |
| ... | ... | ... | ... | ... |
| 13389 | gas pump | valid | val/n03425413/n03425413_17521.JPEG | |
| 13390 | gas pump | valid | val/n03425413/n03425413_20711.JPEG | |
| 13391 | gas pump | valid | val/n03425413/n03425413_19050.JPEG | |
| 13392 | gas pump | valid | val/n03425413/n03425413_13831.JPEG | |
| 13393 | gas pump | valid | val/n03425413/n03425413_1242.JPEG |
The call to head shows the first few rows in the DataFrame. You can see that there are a few metadata columns, as well as the “img” column we added in.
🗂 Selecting data¶
For more information see the user guide section on Data Selection.
When we create an ImageColumn we don’t load the images into memory. Instead, ImageColumn keeps track of all those filepaths we passed in and only loads the images when they are needed.
When we select a row of the ImageColumn, we get an instance FileCell back. A FileCell is an object that holds everything we need to materialize the cell (e.g. the filepath to the image and the loading function), but stops just short of doing so.
img_cell = df["img"][0]
print(f"Indexing the `ImageColumn` returns an object of type: {type(img_cell)}.")
Indexing the `ImageColumn` returns an object of type: <class 'meerkat.columns.deferred.file.FileCell'>.
To actually materialize the image, we simply call the cell.
img = img_cell()
img

We can subselect a batch of images by indexing with a slice. Notice that this returns a smaller DataFrame.
imgs = df["img"][1:4]
print(f"Indexing a slice of the `ImageColumn` returns a: {type(imgs)}.")
imgs
Indexing a slice of the `ImageColumn` returns a: <class 'meerkat.columns.deferred.file.FileColumn'>.
| (FileColumn) | |
|---|---|
| 0 | |
| 1 | |
| 2 |
The whole batch of images can be loaded together by calling the column.
imgs();
One can load multiple rows using any one of following indexing schemes:
Slice indexing: e.g.
column[4:10]Integer array indexing: e.g.
column[[0, 4, 6, 11]]Boolean array indexing: e.g.
column[np.array([True, False, False ..., True, False])]
📎 Aside: ImageColumn under the hood, DeferredColumn.¶
If you check out the implementation of ImageColumn (at meerkat/column/image_column.py), you’ll notice that it’s a super simple subclass of DeferredColumn.
What’s a DeferredColumn?
In meerkat, high-dimensional data types like images and videos are typically stored in a DeferredColumn. A DeferredColumn wraps around another column and lazily applies a function to it’s content as it is indexed. Consider the following example, where we create a simple meerkat column…
col = mk.column([0,1,2])
…and wrap it in a deferred column.
dcol = col.defer(fn=lambda x: x + 10)
dcol[1]() # the function is only called at this point!
Critically, the function inside a lambda column is only called at the time the column is called! This is very useful for columns with large data types that we don’t want to load all into memory at once. For example, we could create a DeferredColumn that lazily loads images…
>>> filepath_col = mk.PandasSeriesColumn(["path/to/image0.jpg", ...])
>>> img_col = filepath.defer(lambda x: load_image(x))
An ImageColumn is a just a DeferredColumn like this one, with a few more bells and whistles!
🛠 Applying operations over the DataFrame.¶
When analyzing data, we often perform operations on each example in our dataset (e.g. compute a model’s prediction on each example, tokenize each sentence, compute a model’s embedding for each example) and store them. The DataFrame makes it easy to perform these operations:
Produce new columns (via
DataFrame.map)Produce new columns and store the columns alongside the original data (via
DataFrame.update)Extract an important subset of the datset (via
DataFrame.filter).
Under the hood, dataloading is multiprocessed so that costly I/O doesn’t bottleneck our computation.
Let’s start by filtering the DataFrame down to the examples in the validation set.
valid_df = df[df["split"] == "valid"]
🫐 Using DataFrame.map to compute average intensity of the blue color channel in the images.¶
To demonstrate the utility of the map operation, we’ll explore the relationship between the “blueness” of an image and the class of the image.
We’ll quantify the “blueness” of each image by simply computing the mean intensity of the blue color channel. This can be accomplished with a simple map operation over the DataFrame:
blue_col = valid_df.map(
lambda img: np.array(img)[:, :, 2].mean(),
num_workers=2
)
# Add the intensities as a new column in the `DataFrame`
valid_df["avg_blue"] = blue_col
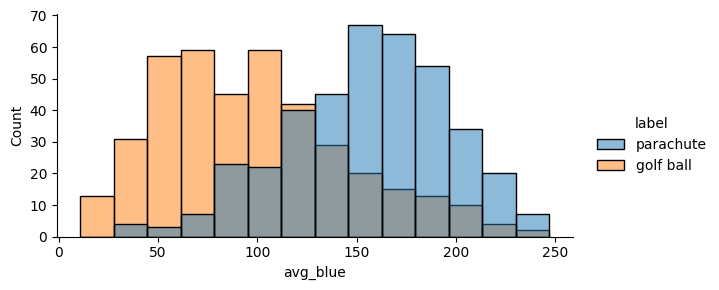
🪂 vs. ⛳️¶
Next, we’ll explore the relationship between blueness and the class label of the image. To do so, we’ll compare the blue intensity distribution of images labeled “parachute” to the distribution of of images labeled “golf ball”.
Using the seaborn plotting package and our DataFrame, this can be accomplished in one line:
## OPTIONAL: this cell requires the seaborn dependency: https://seaborn.pydata.org/installing.html
import seaborn as sns
plot_df = valid_df[np.isin(valid_df["label"], ["golf ball", "parachute"])]
sns.displot(
data=plot_df.to_pandas(),
x="avg_blue",
hue="label",
height=3,
aspect=2
)
/home/runner/work/meerkat/meerkat/meerkat/dataframe.py:901: UserWarning: Could not convert column img of type <class 'meerkat.columns.deferred.file.FileColumn'>, it will be dropped from the output.
warnings.warn(
<seaborn.axisgrid.FacetGrid at 0x7f9c07685be0>
valid_df["img"][int(np.argmax(valid_df["avg_blue"]))]
FileCell(fn=<meerkat.columns.deferred.file.FileLoader object at 0x7f9ca8184b50>)
💾 Writing a DataFrame to disk.¶
Finally, we can write the updated DataFrame to disk for later use.
valid_df.write(os.path.join(dataset_dir, "valid_df"))
valid_df = mk.read(os.path.join(dataset_dir, "valid_df"))

